CytoBrowser
Download the latest release from: https://github.com/MIDA-group/CytoBrowser/releases/latest
Or alternatively from Zenodo via
https://www.npmjs.com/package/cytobrowser
ReadMe, including brief usage example
Rydell C and Lindblad J. "CytoBrowser: a browser-based collaborative annotation platform for whole slide images". F1000Research 2021, 10:226 (https://doi.org/10.12688/f1000research.51916.1)
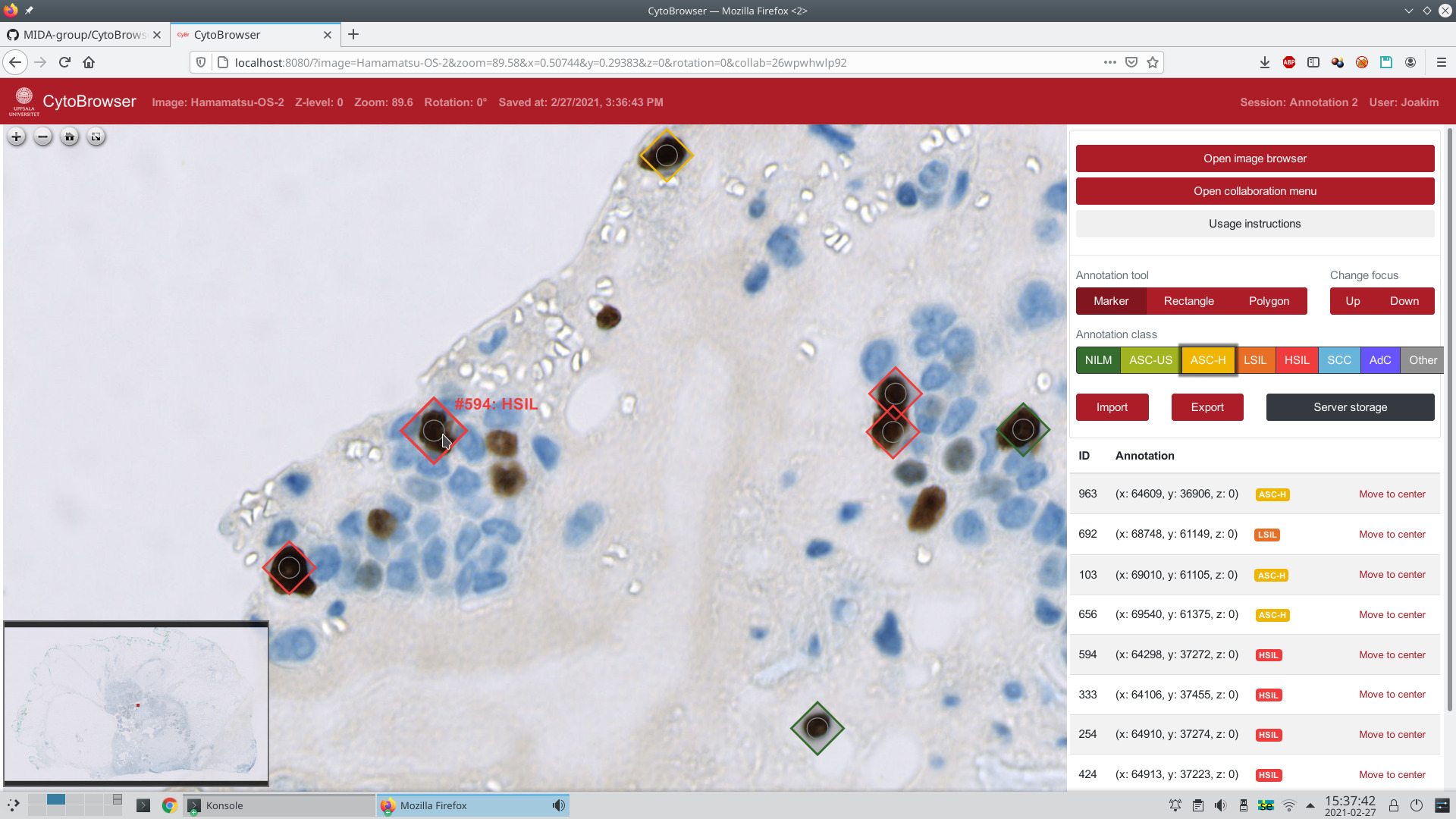
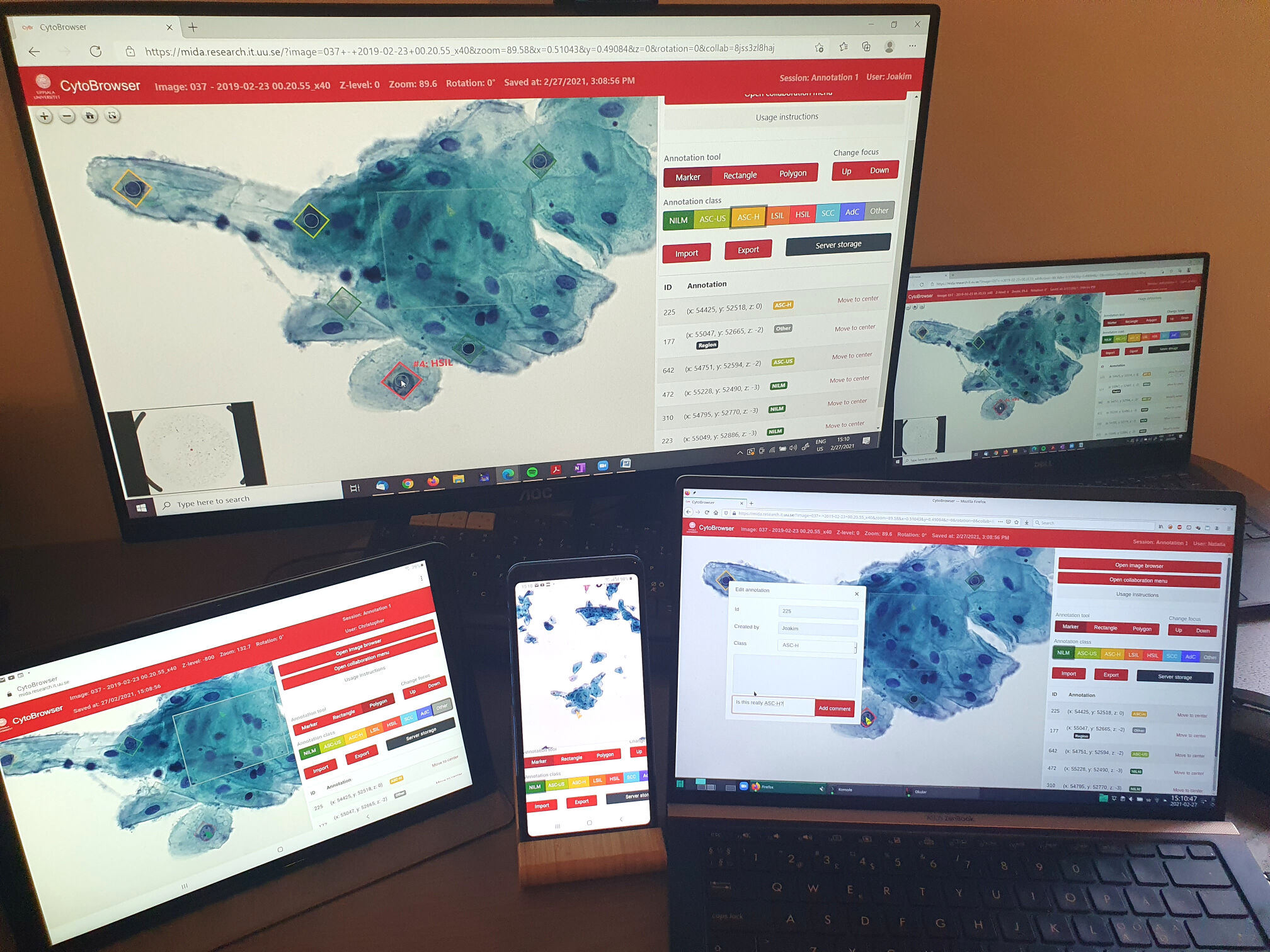
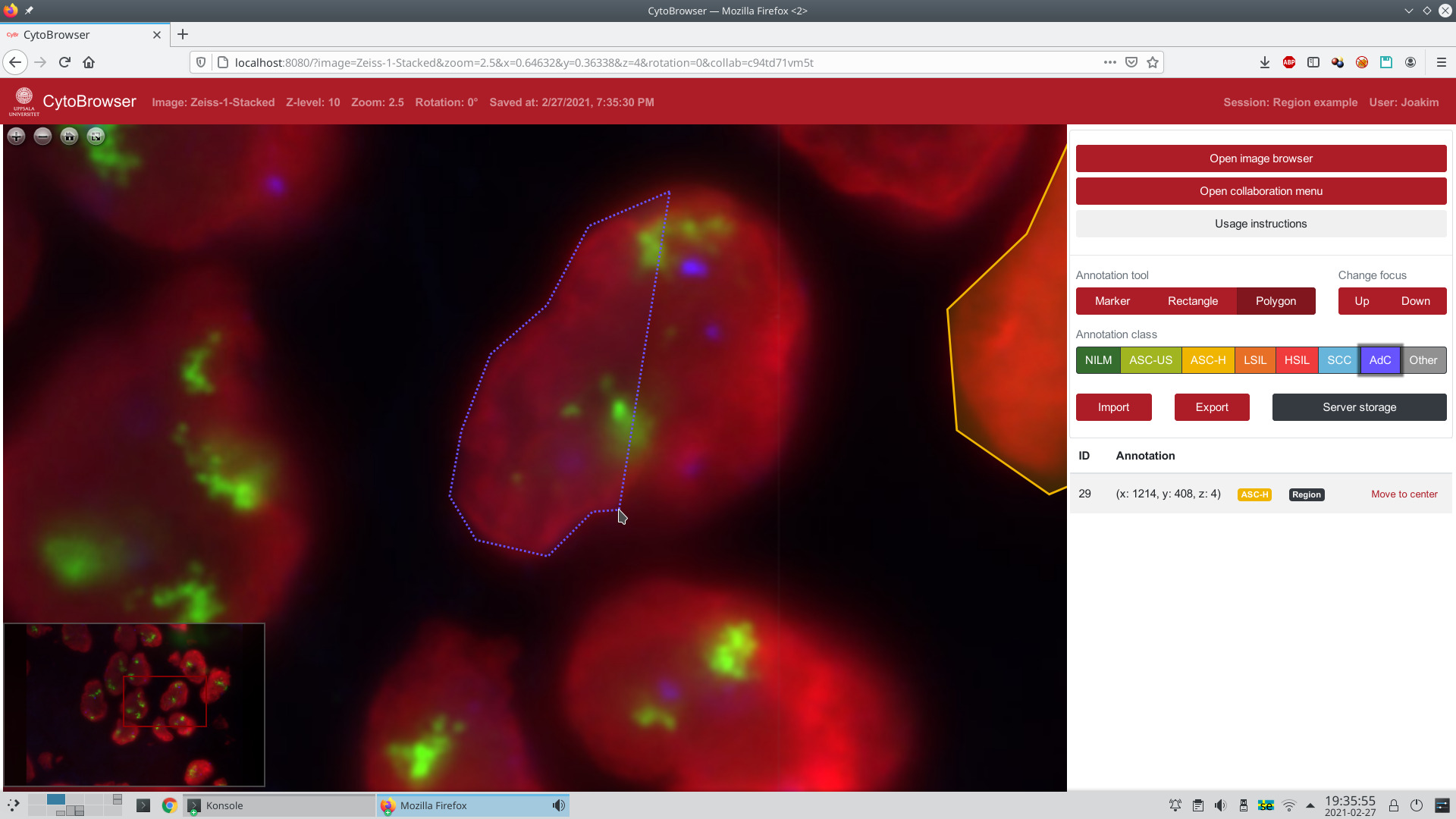
| CytoBrowser is an open-source (GPLv3) JavaScript and Node.js driven environment for fast and accessible collaborative online visualization, assessment, and annotation of very large microscopy images, including, but not limited to, z-stacks (focus stacks) of cytology or histology whole slide images. It provides a web-based viewer for high-resolution zoomable images and facilitates easy remote collaboration, with options for joint-view visualization and simultaneous collaborative annotation of very large datasets. It delivers a combination of functionalities suitable for large scale annotation of whole slide image data. The web browser interface is directly accessible on any modern computer, without need for additional software. By sharing a "session", several remote users can interactively explore and jointly annotate whole slide image data, thereby reaching improved data understanding and annotation quality, effortless project scaling and distribution of resources to/from remote locations, efficient creation of "ground truth" annotations for methods' evaluation and training of machine learning-based approaches, a user-friendly learning environment for medical students, to name a few. Rectangle and polygon region annotations complement point-based annotations, each with a selectable annotation-class as well as free-form text fields. The default setting of CytoBrowser presents an interface for the Bethesda cancer grading system, while other annotation schemes can easily be incorporated. Automatic server side storage of annotations is complemented by JSON-based import/export options facilitating easy interoperability with other tools. |